分子别名(Synonym)
Spike,S1 protein,Spike glycoprotein Subunit1,S glycoprotein Subunit1,Spike protein S1
表达区间及表达系统(Source)
SARS-CoV-2 S1 protein (N501Y), His Tag (S1N-C52Hg) is expressed from human 293 cells (HEK293). It contains AA Val 16 - Arg 685 (Accession # QHD43416.1 (N501Y)). N501Y mutaion was identified in multiple SARS-CoV-2 Variants of Concerns (VOCs), i.e. the Alpha variant (Pango lineage: B.1.1.7; other names: 20I/501Y.V1 or VOC 202012/01), the Beta variant (Pango lineage: B.1.351; other names: 20H/501Y.V2), the Gamma variant (Pango lineage: P.1; other names: 20J/501Y.V3).
Predicted N-terminus: Val 16
Request for sequence
蛋白结构(Molecular Characterization)

This protein carries a polyhistidine tag at the C-terminus
The protein has a calculated MW of 76.9 kDa. The protein migrates as 110-130 kDa under reducing (R) condition (SDS-PAGE) due to glycosylation.
内毒素(Endotoxin)
Less than 1.0 EU per μg by the LAL method.
纯度(Purity)
>90% as determined by SDS-PAGE.
制剂(Formulation)
Lyophilized from 0.22 μm filtered solution in PBS, pH7.4 with trehalose as protectant.
Contact us for customized product form or formulation.
重构方法(Reconstitution)
Please see Certificate of Analysis for specific instructions.
For best performance, we strongly recommend you to follow the reconstitution protocol provided in the CoA.
存储(Storage)
For long term storage, the product should be stored at lyophilized state at -20°C or lower.
Please avoid repeated freeze-thaw cycles.
This product is stable after storage at:
- -20°C to -70°C for 12 months in lyophilized state;
- -70°C for 3 months under sterile conditions after reconstitution.
质量管理控制体系(QMS)
电泳(SDS-PAGE)
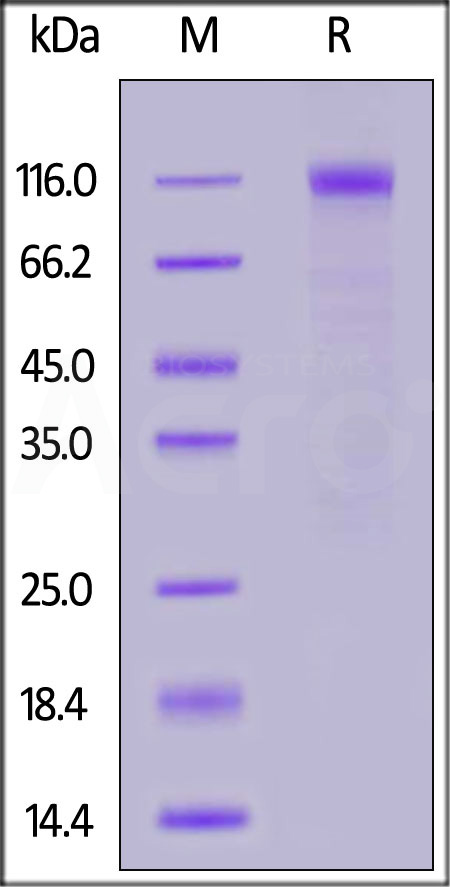
SARS-CoV-2 S1 protein (N501Y), His Tag on SDS-PAGE under reducing (R) condition. The gel was stained with Coomassie Blue. The purity of the protein is greater than 90%.
活性(Bioactivity)-ELISA
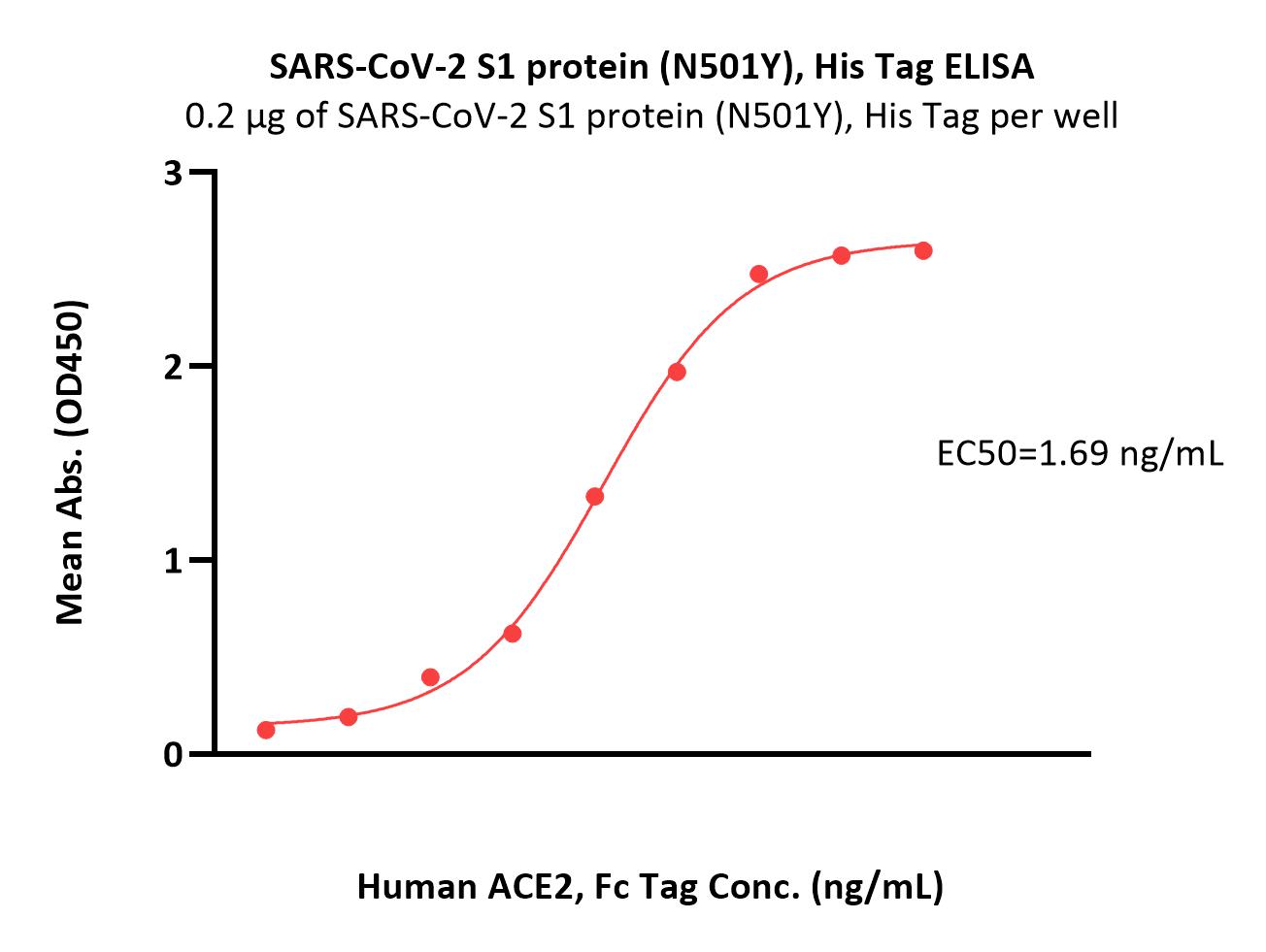
Immobilized SARS-CoV-2 S1 protein (N501Y), His Tag (Cat. No. S1N-C52Hg) at 2 μg/mL (100 μL/well) can bind Human ACE2, Fc Tag (Cat. No. AC2-H5257) with a linear range of 0.1-3 ng/mL (QC tested).
Protocol
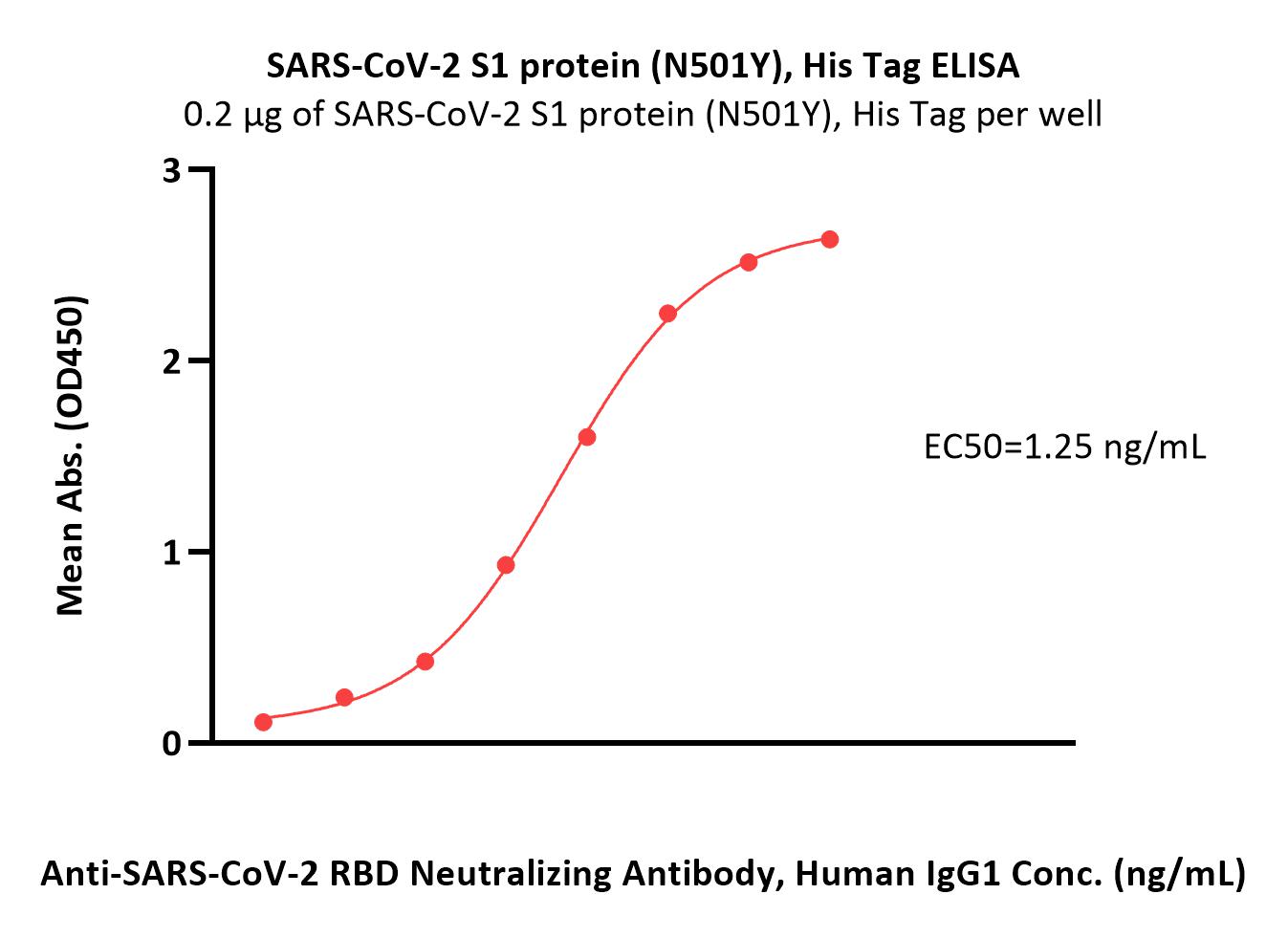
Immobilized SARS-CoV-2 S1 protein (N501Y), His Tag (Cat. No. S1N-C52Hg) at 2 μg/mL (100 μL/well) can bind Anti-SARS-CoV-2 RBD Neutralizing Antibody, Human IgG1 (Cat. No. SAD-S35) with a linear range of 0.1-3 ng/mL (Routinely tested).
Protocol
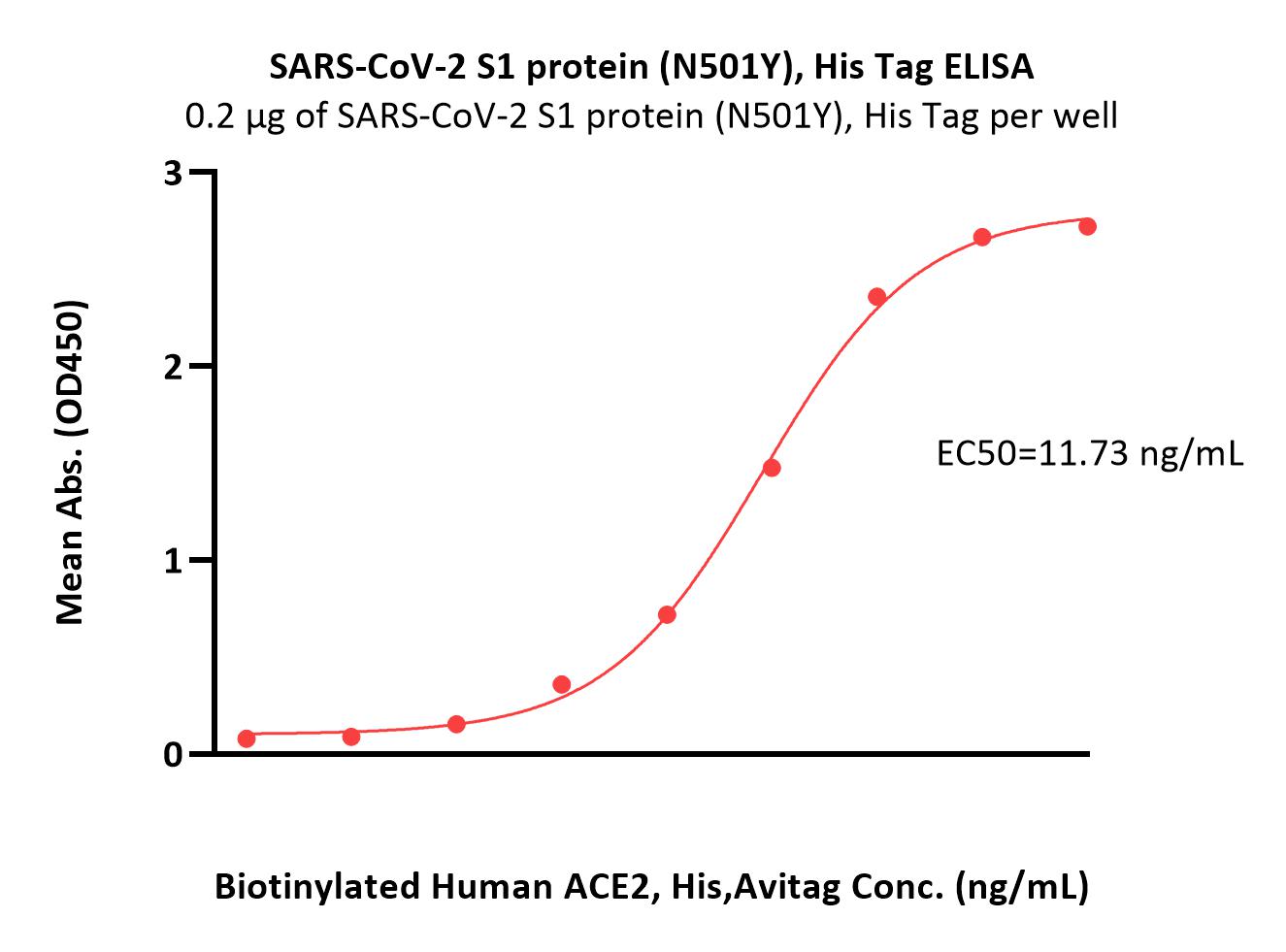
Immobilized SARS-CoV-2 S1 protein (N501Y), His Tag (Cat. No. S1N-C52Hg) at 2 μg/mL (100 μL/well) can bind Biotinylated Human ACE2, His,Avitag (Cat. No. AC2-H82E6) with a linear range of 0.8-25 ng/mL (Routinely tested).
Protocol
背景(Background)
It's been reported that Coronavirus can infect the human respiratory epithelial cells through interaction with the human ACE2 receptor. The spike protein is a large type I transmembrane protein containing two subunits, S1 and S2. S1 mainly contains a receptor binding domain (RBD), which is responsible for recognizing the cell surface receptor. S2 contains basic elements needed for the membrane fusion.The S protein plays key parts in the induction of neutralizing-antibody and T-cell responses, as well as protective immunity.























































 膜杰作
膜杰作 Star Staining
Star Staining