Flow cytometric analysis of the SARS coronavirus 2 antibodies in human plasmaFang, Shrestha, Beland
Sci Rep (2025) 15 (1), 10300
Abstract: COVID-19 is an infectious disease caused by the severe acute respiratory syndrome coronavirus (SARS-CoV-2). Anti-SARS-CoV-2 antibodies can provide information on patient immunity, identify asymptomatic patients, and track the spread of COVID-19. Efforts have been made to develop methods to detect anti-SARS-CoV-2 antibodies in humans. Here, we describe a flow cytometric assay for the simultaneous detection of anti-SARS-CoV-2 IgG and IgM in human plasma. To assess the antibody response against the different SARS-CoV-2 structural proteins, five viral recombinant proteins, including spike protein subunit 1 (S1), N-terminal domain of S1 (S1A), spike receptor-binding domain (RBD), spike protein subunit 2 (S2), and nucleocapsid protein (N), were generated. A comparison of the antibody profiles detected by the assay with plasma from 100 healthy blood donors collected prior to the COVID-19 pandemic and plasma from 100 virologically confirmed COVID-19 patients demonstrated a clear discrimination between the two groups. Among the COVID-19 patients, the antibody responses for the viral proteins, as determined by their prevalence, were anti-RBD IgG = anti-N IgG > anti-S1 IgG > anti-S1A IgG > anti-S2 IgG, and anti-RBD IgM > anti-S1 IgM > anti-N IgM > anti-S2 IgM. The prevalence of anti-SARS-CoV-2 IgG and IgM was not associated with sex, age, race, days after the onset of symptoms, or severity of illness, except for a higher prevalence of anti-S2 IgG being observed in men than in women. The levels of anti-RBD IgG were higher in patients 65 years and older and in patients who had severe symptoms. Similarly, patients who had severe symptoms exhibited higher levels of anti-S1 and anti-S1A IgG than patients who had mild or moderate symptoms. The levels of anti-RBD IgM tended to be higher in men but did not differ among age, race, days after the onset of symptoms, or severity of illness. Our study indicates that the flow cytometric assay, especially using RBD as target antigen, can be used to detect simultaneously anti-SARS-CoV-2 IgG and IgM antibodies in human plasma.© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Alveolar epithelial type 2 cell specific loss of IGFBP2 activates inflammation in COVID-19Pujadas, Chin, Sankpal
et alRespir Res (2025) 26 (1), 111
Abstract: The coronavirus disease 2019 (COVID-19) global pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, our understanding of SARS-CoV-2-induced inflammation in alveolar epithelial cells remains very limited. The contributions of intracellular insulin-like growth factor binding protein-2 (IGFBP2) to SARS-CoV-2 pathogenesis are also unclear. In this study, we have uncovered a critical role for IGFBP2, specifically in alveolar epithelial type 2 cells (AEC2), in the immunopathogenesis of COVID-19. Using bulk RNA sequencing, we show that IGFBP2 mRNA expression is significantly downregulated in primary AEC2 cells isolated from fibrotic lung regions from patients with COVID-19-acute respiratory distress syndrome (ARDS) compared to those with idiopathic pulmonary fibrosis (IPF) alone or IPF with a history of COVID-19. Using multicolor immunohistochemistry, we demonstrated that IGFBP2 and its selective ligands IGF1 and IGF2 were significantly reduced in AEC2 cells from patients with COVID-ARDS, IPF alone, or IPF with COVID history than in those from age-matched donor controls. Further, we demonstrated that lentiviral expression of Igfbp2 significantly reduced mRNA expression of proinflammatory cytokines-Tnf-α, Il1β, Il6, Stat3, Stat6 and chemokine receptors-Ccr2 and Ccr5-in mouse lung epithelial cells challenged with SARS-CoV-2 spike protein injury (S2; 500 ng/mL). Finally, we demonstrated higher levels of cytokines-TNF-α; IL-6 and chemokine receptor-CCR5 in AEC2 cells from COVID-ARDS patients compared to the IPF alone and the IPF with COVID history patients. Altogether, these data suggest that anti-inflammatory properties of IGFBP2 in AEC2 cells and its localized delivery may serve as potential therapeutic strategy for patients with COVID-19.© 2025. The Author(s).
The fusion peptide of the spike protein S2 domain may be a mimetic analog of β-coronaviruses and serve as a novel virus-host membrane fusion inhibitorSafiriyu, Hussain, Dewangan
et alAntiviral Res (2025) 237, 106144
Abstract: Coronavirus has garnered more attention recently, particularly in the aftermath of the 2019 pandemic. The β genus of the coronavirus family has demonstrated a significant threat to humanity. Current mitigation strategies involve the development of vaccines and repurposing drugs for symptomatic management of coronavirus infection, specifically SARS-Cov 2. Fusion inhibitors that are available as antiviral drugs for coronavirus have targeted the heptad repeat (HR) 1 and 2 in the S2 domain of the spike protein. The current study identified a fusion peptide (FP) upstream of HR1 as a potential target for developing membrane fusion inhibitors, and mimetic peptides analogous to the FP segment were tested for antiviral activity. Four mimetic fusion peptides (MFPs) (RSA59PP (MFP633), RSA59P (MFP634), RSMHV2P (MFP635), and RSMHV2PP (MFP636)) that are analogous to the FP of murine β coronavirus mouse hepatitis virus (MHV), MHV-A59/RSA59 (PP) and MHV-2/RSMHV2 (P) with central proline mutations, were tested. Results show the ability of MFPs to reduce cell-to-cell fusion and viral replication in vitro. MFP633, which contains a central double proline, exhibited the most potent inhibitory effect in spike protein-mediated membrane fusion assays. Biophysical experiments also demonstrated the strongest interactions between double-proline containing MFPs (MFP633 and MFP636) with biomimetic liposomes. In vivo studies using a liposome-mediated delivery system in mice confirmed the antiviral activity of MFP633. These findings suggest that targeting FPs could develop effective fusion inhibitors against coronaviruses. MFPs act on the host cell membrane by competing with the viral FP during the early stage of host-viral membrane fusion events. MFP633 is a promising peptide drug candidate that warrants future examination to assess whether this and other dual-proline containing peptides may exert similar anti-viral effects in other coronaviruses with conserved FP structures.Copyright © 2025. Published by Elsevier B.V.
SARS CoV-2 spike adopts distinct conformational ensembles in situGramm, Braet, Srinivasu
et albioRxiv (2025)
Abstract: Engineered recombinant Spike (S) has been invaluable for determining S structure and dynamics and is the basis for the design of most prevalent vaccines. While these vaccines have been highly efficacious for short-term protection from infection, protection waned with the emergence of variants (alpha through omicron). Here we report differences in conformational dynamics between native, membrane-embedded full-length S and recombinant S. Our virus-like particle (VLP) model mimics the native SARS CoV-2 virion by displaying S assembled with auxiliary E, M, and N proteins in a native membrane environment that captures the entirety of quaternary interactions mediated by S. Display of S on VLP obviates the requirement for stabilizing modifications that have been engineered into recombinant S for enhanced expression and solubility. Amide hydrogen/deuterium exchange mass spectrometry (HDXMS) reveals altered interprotomer contacts in VLP S trimers attributable to the presence of auxiliary proteins, membrane anchoring, and lack of engineered modifications. Our results reveal decreased dynamics in the S2 subunit and at sites spanning interprotomer contacts in VLP S with minimal differences in the N-terminal domain (NTD) and receptor binding domain (RBD). This carries implications for display of epitopes beyond NTD and RBD. In summary, despite affording efficient structural characterization, recombinant S distorts the intrinsic conformational ensemble of native S displayed on the virus surface.

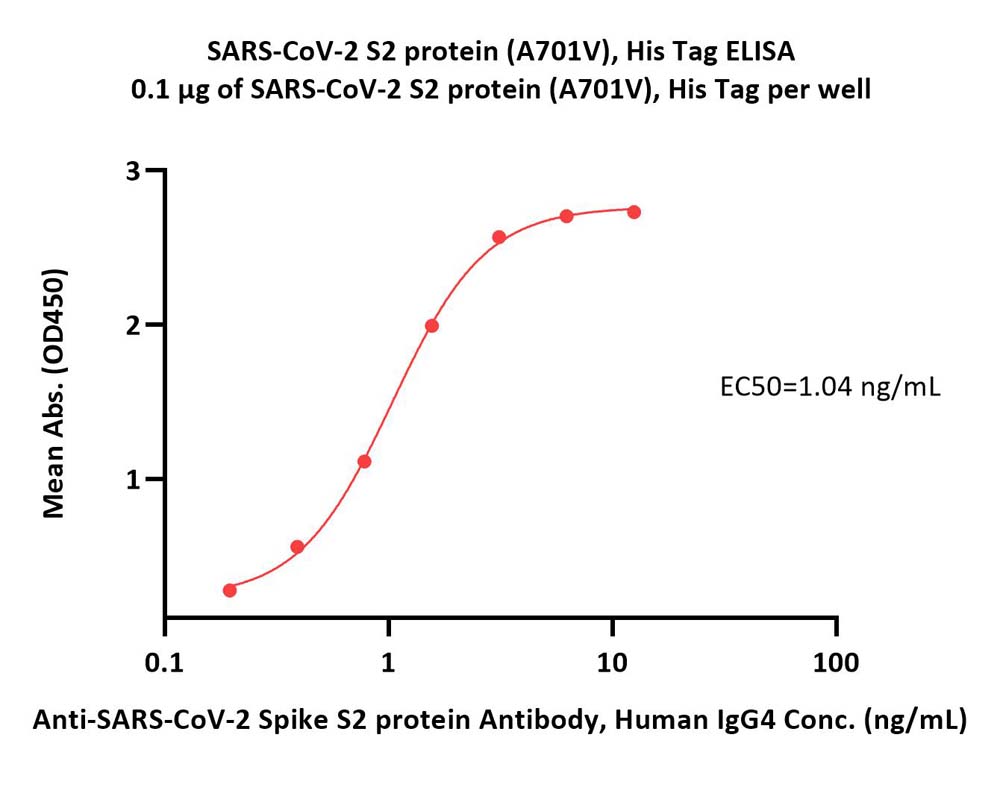
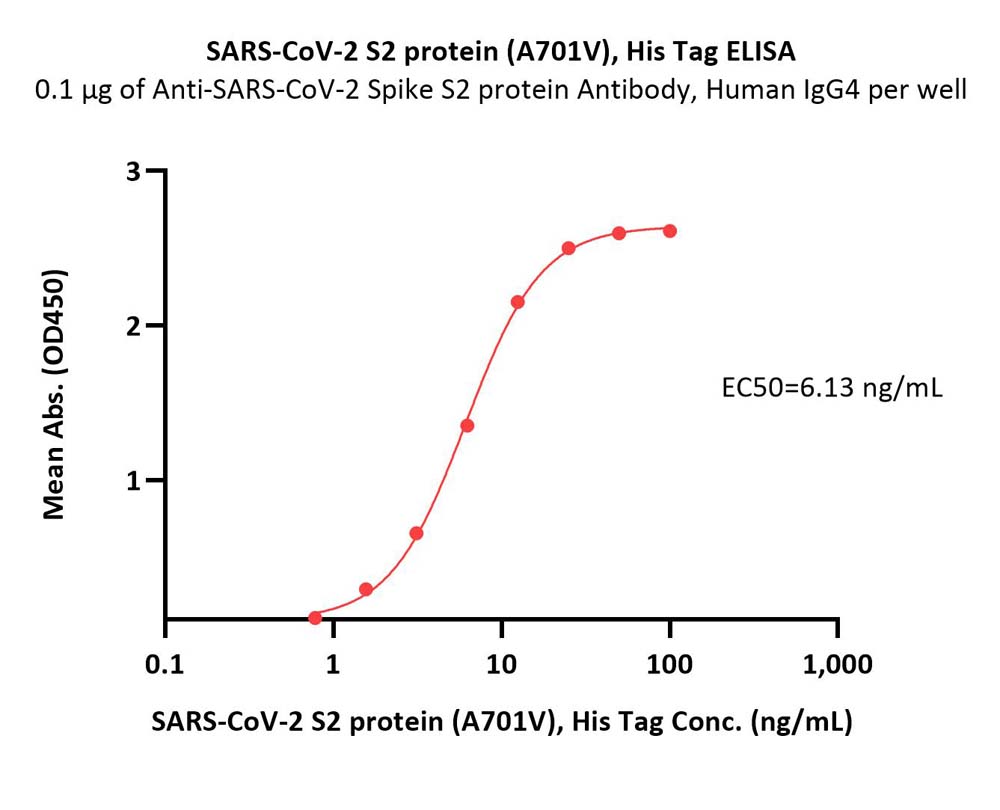























































 膜杰作
膜杰作 Star Staining
Star Staining















