| Items | Size (2mg) | Size(5mg X 2) |
| Particle size | 2 μm | 2 μm |
| Physical appearance | Powder mixture | Powder mixture |
| Amount of Coupled Protein | ≈286 pmol (40 μg) SARS-CoV-2 Spike Trimer (P.1)/mg beads | ≈286 pmol (40 μg) SARS-CoV-2 Spike Trimer (P.1)/mg beads |
| Binding Capacity | >242 pmol (36 μg) Anti-SARS-CoV-2 Spike RBD Neutralizing Antibody/mg beads | >242 pmol (36 μg) Anti-SARS-CoV-2 Spike RBD Neutralizing Antibody/mg beads |
| Formulation | PBS, pH7.4, with 10% Trehalose | PBS, pH7.4, with 10% Trehalose |
| Reconstitution | 2 mL sterile deionized water (1 mg beads/mL) | 5 mL sterile deionized water (1 mg beads/mL) |
背景(Background)
The SARS-CoV-2 Spike Trimer (P.1) Coupled Magnetic Beads is produced by coupling biotinylated SARS-CoV-2 spike trimer to streptavidin-conjugated magnetic beads. The spike proteins coupled to the beads contain 12 mutations (L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F) identified in the Brazilian variant (known as P.1). The pre-coupled beads are ready to use for capturing anti-SARS-CoV-2 antibody or ACE2 protein from your sample with high specificity.
表达区间及表达系统(Source)
SARS-CoV-2 Spike Trimer (P.1) Protein is expressed from human 293 cells (HEK293). It contains AA Val 16 - Pro 1213(Accession # QHD43416.1 (L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F, R683A, R685A,F817P, A892P, A899P, A942P, K986P, V987P)).
应用说明(Application)
This product is intended for immunocapture, biopanning. This is a non-sterile product.
重构方法(Reconstitution)
See Certificate of Analysis (CoA) for detailed instruction.
存储(Storage)
Upon receipt, please store the Beads at -20°C. The shelf life is 1 year at -20 °C.
Please avoid more than 3 freeze-thaw cycles once reconstitution, immediate use after reconstitution is highly recommended.
原理(Assay Principles)
Antibody Purification: 1. Resuspend the lyophilized beads by adding the buffer of choice. 2. Add analyte to the suspension, mix and incubate to enable specific binding of the beads and the target protein. 3. Magnetize beads, remove supernatant, and wash unbound protein fractions to capture target protein-bound beads. 4. Wash, magnetize the beads and collect purified target protein for use in downstream applications.
The magnetic beads technology makes use of the easy and efficient collection of beads in magnetic field to facilitate antibody purification in a simple workflow of “bind-wash-elute”. In contrast to common separation techniques, this method does not require columns or centrifugation, and is therefore ideal in high-throughput applications.
制剂(Formulation)
Please contact us for detailed information.
Contact us for customized product form or formulation.
质量管理控制体系(QMS)
典型数据-Typical Data
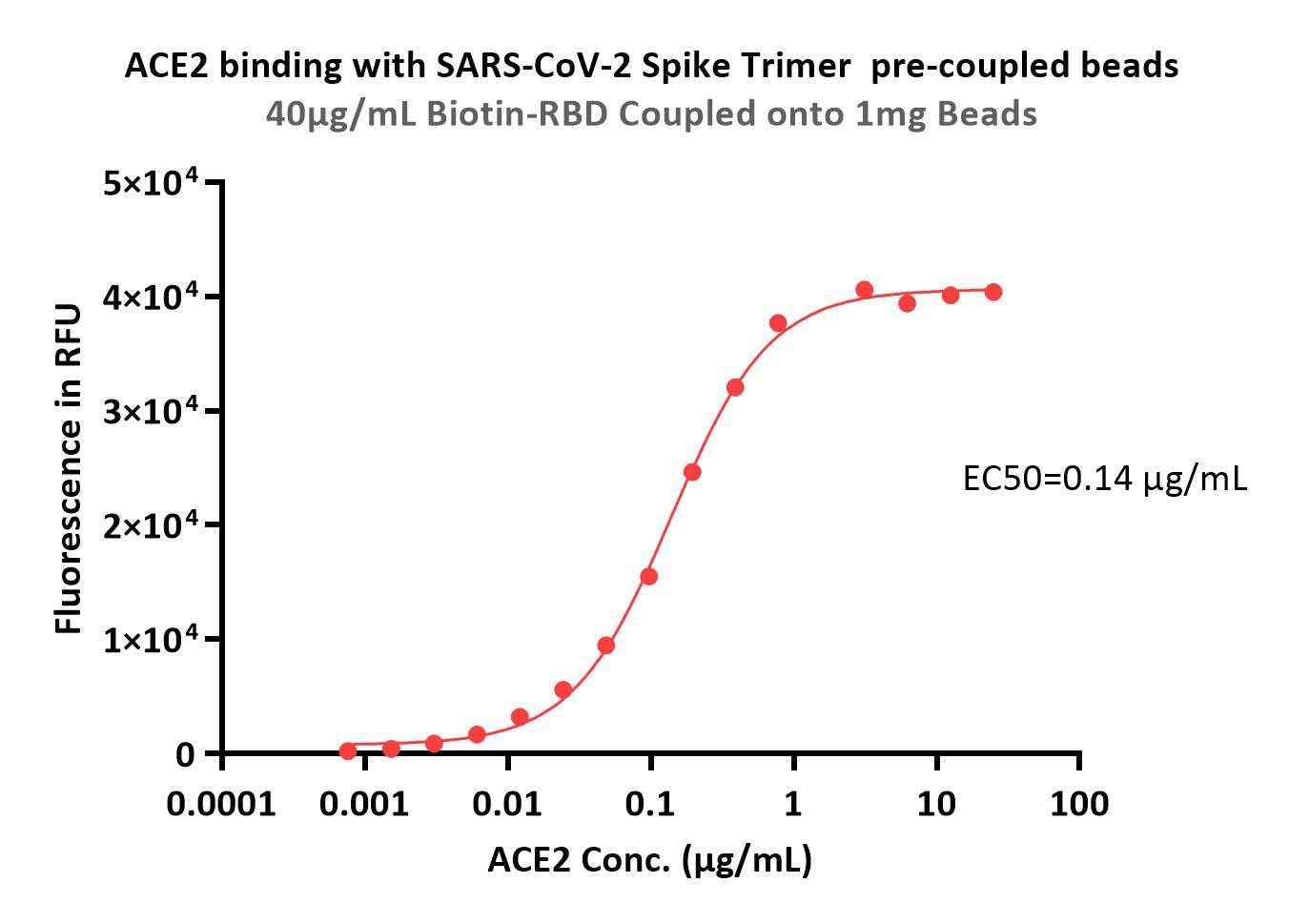
Immobilized 40 μg SARS-CoV-2 S protein trimer to 1mg Beads can bind the ACE2 with an EC50 of 0.14μg/mL (QC tested).
Protocol
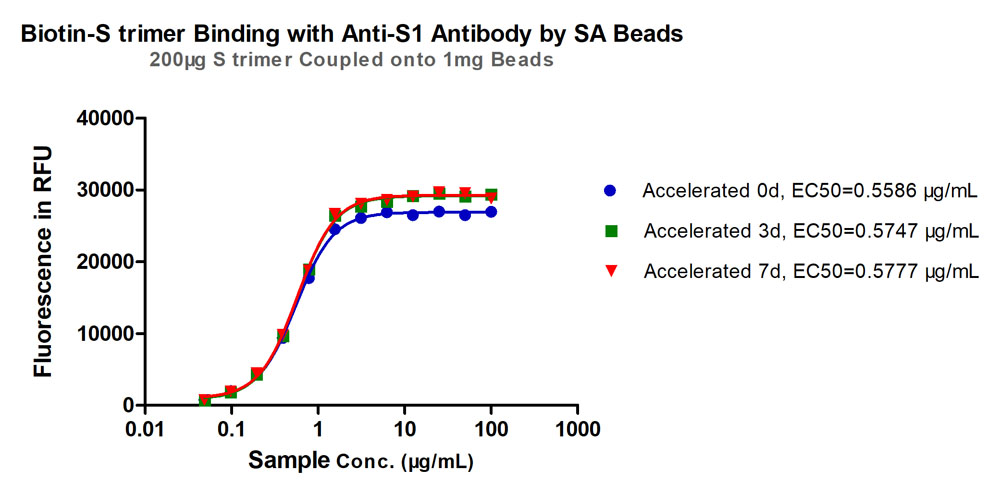
Accelerated stability test. After placing the lyophilized beads at 37°C for 7 days, binding activity between the S SARS-CoV-2 Spike Trimer (P.1) Coupled Magnetic Beads (Cat.No. MBS-K031) and anti-SARS-CoV-2 Spike S1 antibody showed little deviation from the unaccelerated sample (%RSD<5%). Data were measured on day 0, 3, 7 respectively.
Protocol
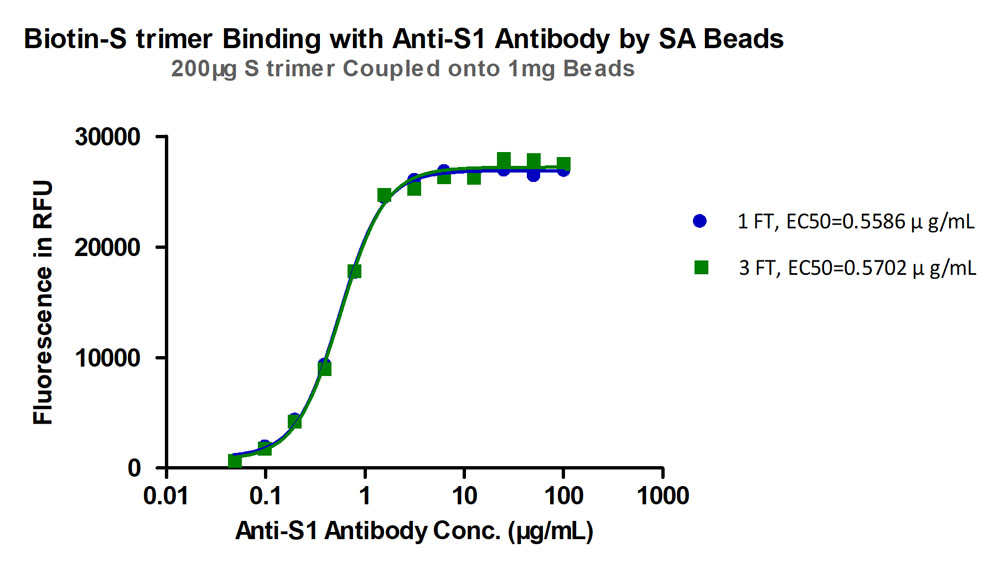
Freeze-thaw stability test. After different freeze-thaw cycles, binding activity between SARS-CoV-2 Spike Trimer (P.1) Coupled Magnetic Beads (Cat.No. MBS-K031) and anti-SARS-CoV-2 Spike S1 antibody showed little deviation from the unfree-thaw sample (%RSD<5%). Three freeze-thaw cycles were performed.
Protocol























































 膜杰作
膜杰作 Star Staining
Star Staining







 & Cat.No.
& Cat.No. 







