EGFR-mediated local invasiveness and response to Cetuximab in head and neck cancerZhou, He, Zhao
et alMol Cancer (2025) 24 (1), 94
Abstract: Recurrent/metastatic head and neck squamous cell carcinoma (R/M-HNSCC) is a severe, frequently lethal condition. Oncogene addiction to epidermal growth factor receptor (EGFR) is a hallmark of HNSCC, but the clinical efficacy of EGFR-targeted therapies remains low. Understanding molecular networks governing EGFR-driven progression is paramount to the exploration of (co)-treatment targets and predictive markers.We performed function-based mapping of differentially expressed genes in EGFR-mediated local invasion (fDEGs) using photoconvertible tracers and RNA-sequencing (RNA-seq) in a cellular 3D-model.Upon alignment with public single-cell RNA-seq (scRNA-seq) datasets and HNSCC-specific regulons, a gene regulatory network of local invasion (invGRN) was inferred from gene expression data, which was overrepresented in budding tumors. InvGRN comprises the central hubs inhibin subunit beta alpha (INHBA) and snail family transcriptional repressor 2 (SNAI2), and druggable fDEGs integrin subunit beta 4 (ITGB4), laminin 5 (LAMB3/LAMC2), and sphingosine kinase 1 (SPHK1). Blockade of INHBA repressed local invasion and was reverted by activin A, laminin 5, and sphingosine-1-phosphate, demonstrating a functional interconnectivity of the invGRN. Epithelial-to-mesenchymal transition (EMT) of malignant cells and the invGRN are induced by newly defined EGFR-activity subtypes with prognostic value that are promoted by amphiregulin (AREG) and epiregulin (EREG). Importantly, co-inhibition of SPHK1 showed synthetic effects on Cetuximab-mediated invasion blockade and high expression of selected fDEGs was associated with response to Cetuximab in patient-derived xenotransplantation (PDX) and R/M-HNSCC patients.We describe an actionable network of EGFR-mediated local invasion and define druggable effectors with predictive potential regarding the response of R/M-HNSCC to Cetuximab.© 2025. The Author(s).
Structural insights on perlecan and Schwartz-Jampel syndromeSohail, Koski, Ruddock
Matrix Biol (2025)
Abstract: Perlecan is an essential multi-domain, disulfide bond rich basement membrane protein. Mutations in perlecan cause Schwartz-Jampel syndrome and dyssegmental dysplasia. While there has been a large body of experimental work reported on perlecan, there is only minimal structural information available to date. There is no prior structural data for region 3 of perlecan in which some Schwartz-Jampel syndrome causing point mutations have been reported. Here, we produce constructs of the disulfide rich region 3 of perlecan along with five mutations previously reported to cause Schwatz-Jampel syndrome. Four of the mutations resulted in decreased yields and thermal stability compared to the wild-type protein. In contrast, the P1019L mutation was produced in good yields and showed higher thermal stability than the wild-type protein. The crystal structures for both the wild-type and P1019L mutation were solved. As expected, both showed laminin IV-like and laminin-type EGF-like domains, with the P1019L mutation resulting in only a minor conformational change in a loop region and no significant changes in regular secondary or tertiary structure.Copyright © 2025. Published by Elsevier B.V.
Self-organization of mouse embryonic stem cells into reproducible pre-gastrulation embryo models via CRISPRa programmingLodewijk, Kozuki, Han
et alCell Stem Cell (2025)
Abstract: Embryonic stem cells (ESCs) can self-organize into structures with spatial and molecular similarities to natural embryos. During development, embryonic and extraembryonic cells differentiate through activation of endogenous regulatory elements while co-developing via cell-cell interactions. However, engineering regulatory elements to self-organize ESCs into embryo models remains underexplored. Here, we demonstrate that CRISPR activation (CRISPRa) of two regulatory elements near Gata6 and Cdx2 generates embryonic patterns resembling pre-gastrulation mouse embryos. Live single-cell imaging revealed that self-patterning occurs through orchestrated collective movement driven by cell-intrinsic fate induction. In 3D, CRISPRa-programmed embryo models (CPEMs) exhibit morphological and transcriptomic similarity to pre-gastrulation mouse embryos. CPEMs allow versatile perturbations, including dual Cdx2-Elf5 activation to enhance trophoblast differentiation and lineage-specific activation of laminin and matrix metalloproteinases, uncovering their roles in basement membrane remodeling and embryo model morphology. Our findings demonstrate that minimal intrinsic epigenome editing can self-organize ESCs into programmable pre-gastrulation embryo models with robust lineage-specific perturbation capabilities.Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
A hollow fiber membrane-based liver organoid-on-a-chip model for examining drug metabolism and transportMyszczyszyn, Münch, Lehmann
et alBiofabrication (2025)
Abstract: Liver-on-a-chip models predictive for both metabolism, and blood and canalicular transport of drug candidates in humans are lacking. Here, we established a bioengineered and 3Rs-complied (animal component-free) hepatocyte-like millifluidic system based on 3D hollow fiber membranes (HFMs), recombinant human laminin 332 coating and adult human stem cell-derived organoids. Organoid fragments formed polarized and tight monolayers on HFMs with improved hepatocyte-like maturation, as compared to standard 3D organoid cultures in Matrigel from matched donors. Gene expression profiling and immunofluorescence revealed that hepatocyte-like monolayers expressed a broad panel of phase I (e.g., CYP3A4, CYP2D6, CYP2C9) and II (e.g., UGTs, SULTs) drug-metabolizing enzymes and drug transporters (e.g., MDR1, MRP3, OATP1B3). Moreover, statically cultured monolayers displayed phase I and II metabolism of a cocktail of six relevant compounds, including midazolam and 7-hydroxycoumarin. We also demonstrated the disposition of midazolam in the basal/blood-like circulation and apical/canalicular-like compartment of the millifluidic chip. Finally, we studied the bioavailability of midazolam and coumarin on-a-chip in combination with a small intestine-like system. In conclusion, we generated a proof-of-concept liver organoid-on-a-chip model for examining metabolism and transport of drugs, which can be further developed to predict PK/ADME profiles in humans.Creative Commons Attribution license.
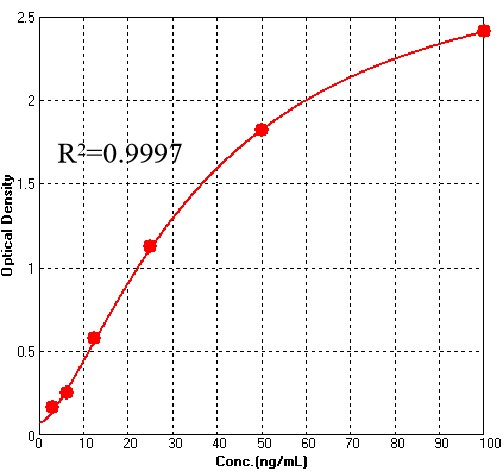
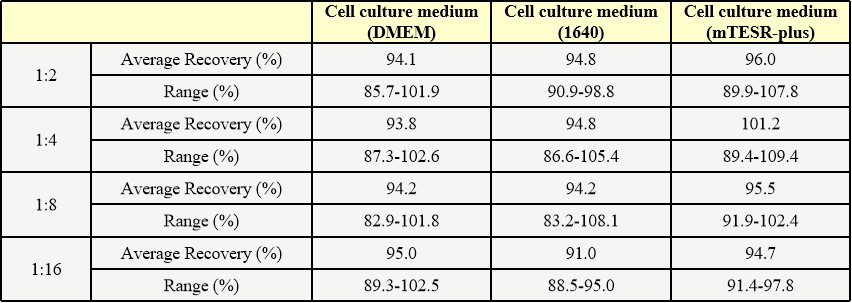
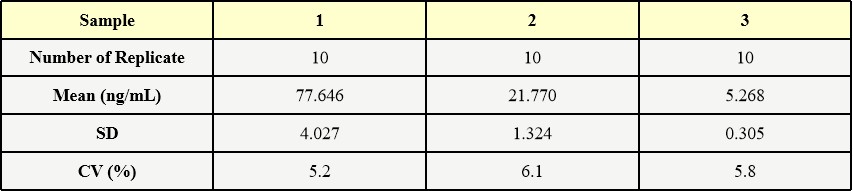
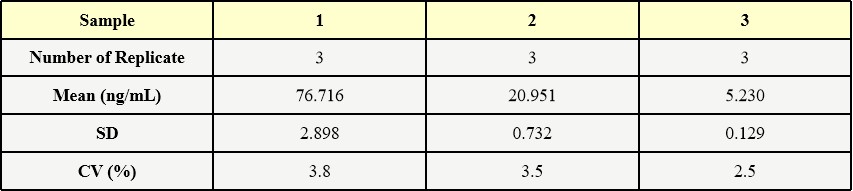























































 膜杰作
膜杰作 Star Staining
Star Staining















