Source
GENIUS™Nuclease is a recombinant form of Serratia marcescens extracellular endonuclease produced in Escherichia coli cells using a proprietary process at ACRObiosystems. GENIUS™Nuclease is a homodimer with monomer molecular masses about 30 kDa. Two disulfide bonds found in the nuclease are crucial to its activity and stability. The enzyme is a non-specific nuclease with high specific activity, which degrades both single- and double-stranded nucleic acids in any form ( single stranded, double stranded, linear, circular and supercoiled). It hydrolyzes internal phosphodiester bonds present between the nucleotides to 5‘- phosphorylated oligonucleotides of 3-5 bases in length.
Application
Its high intrinsic activity and broad substrate tolerance make the endonuclease an ideal tool in a variety of biotechnological and pharmaceutical applications: removal of nucleic acid from protein samples ( Elimination of nucleic acids from recombinant proteins; Purification of protein fragments from inclusion bodies; Sample preparation in western blotting or two- dimensional gel electrophoresis) ; Viscosity reduction in protein extracts.
Operating conditions
GENIUS™Nuclease is functional between pH 6 and 10 (optimal at pH8 - 8.5) , and from 0℃ to 42 ℃ (optimal at 35 ℃ - 42 ℃). Mg2+ (1-2 mM) is required for enzyme activity.
1 mM EDTA reduced the activity by 30% in the presence of 1 mM MgCl2; 0.1 M EDTA eliminated all enzyme activity. In the presence of 1 mM MgCl2, enzyme levels were reduced 75% by 0.1 M CaCl2 or 1 M NaCl. Under standard assay conditions, 1 mM iodoacetate had no effect on the enzymatic rate, whereas 1 mM mercaptoethanol and maleic acid reduced the activity by only 5 to 10%. 10 mM p- Chloromercurybenzoate completely inactivates the enzyme, while 0.64 M beta-mercaptoethanol in the presence of 2 M urea causes only partial inactivation of the enzyme. 4 or 7 M Urea increases the enzyme activity.
Removal of GENIUS™Nuclease
GENIUS™Nuclease contain no “Tag” and used in downstream processing can be removed by various purification methods according to the purification strategy for the target protein.
Formulation
Lyophilized in Tris HCl, pH 8.0, MgCl2, and NaCl.
Reconstitution
See Certificate of Analysis for reconstitution instructions and specific concentrations.
Purity
>95 % as determined by SDS-PAGE reduced GENIUS™Nuclease.
Enzyme Activity
>250U/μL
Activity Assay Procedure
1. Reagents and solutions preparation
Reaction buffer*:
50 mM Tris-HCl, 1 mM MgCl2, pH 8.0 ( * In the case of extensive dilution before use, carrier protein such as 0.1 mg/ml HSA or BSA is generally recommended to avoid any enzyme loss from surface adsorption)
DNA Substrate:
1 mg/ml salmon sperm DNA is dissolved overnight at 4 ℃, in reaction buffer, and is then sonicated on ice to obtain a homogenous solution.
Enzyme:
Different dilution of nuclease with reaction buffer.
Stop reagent:
Trichloroacetic acid (TCA)
2. Standard curve establishment
400 μl substrate + 100 μl enzyme of known activity = 500 μl mixture
- Incubate the mixture at 37℃ for 30 min.
- Stop the reaction by addition of 400 μl cold TCA and incubate on ice for 10 min.
- Centrifuge at 8500g for 5 min.
- Measure the absorbance of supernatant at 260 nm.
- Lot a standard curve with nuclease of known activities for each set of measurements.
3. Measurement of activity
The activity of any unknown nuclease can be determined from a single measurement by means of the standard curve. The specific activity of GENIUS™Nuclease is >1.0 x 10e6 unit/mg protein.
Activity
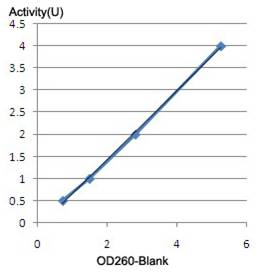
Standard curve of nuclease activity for GENIUS™Nuclease.
Unit Definition
One unit will digest sonicated salmon sperm DNA to acid-soluble oligonucleotides equivalent to a ΔA260 of 1.0 in 30 min at pH 8.0 at 37 ℃, which corresponds approximately to complete digestion of 37 μg DNA. Note that 1 KU=1000 units.
Storage
Avoid repeated freeze-thaw cycles.
This product is stable after storage at:
In lyophilized state for 1 year (-20oC); After reconstitution under sterile conditions for 3 months (-70oC).
Notice: We updated the brand name from Benz™Nuclease to GENIUS™Nuclease. The products are the same and the only change is the brand name in the product name and the label.
电泳(SDS-PAGE)
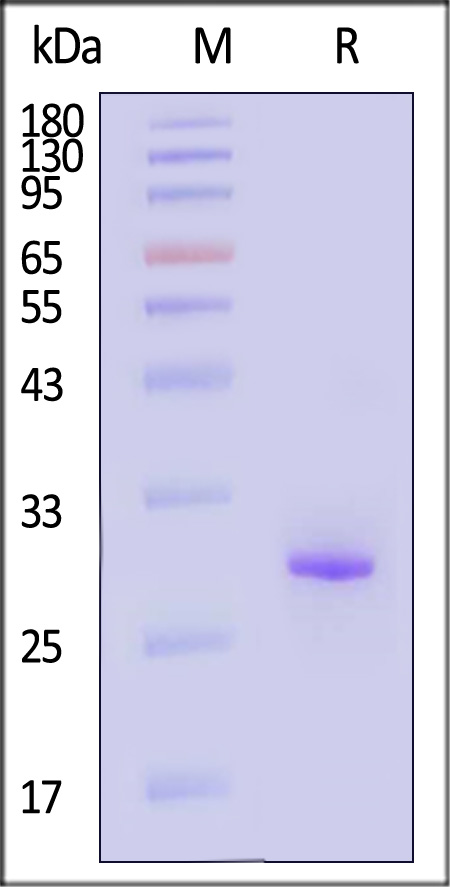
The purity of GENIUS™Nuclease was determined by SDS-PAGE reduced and staining overnight with Coomassie Blue.
SEC-HPLC
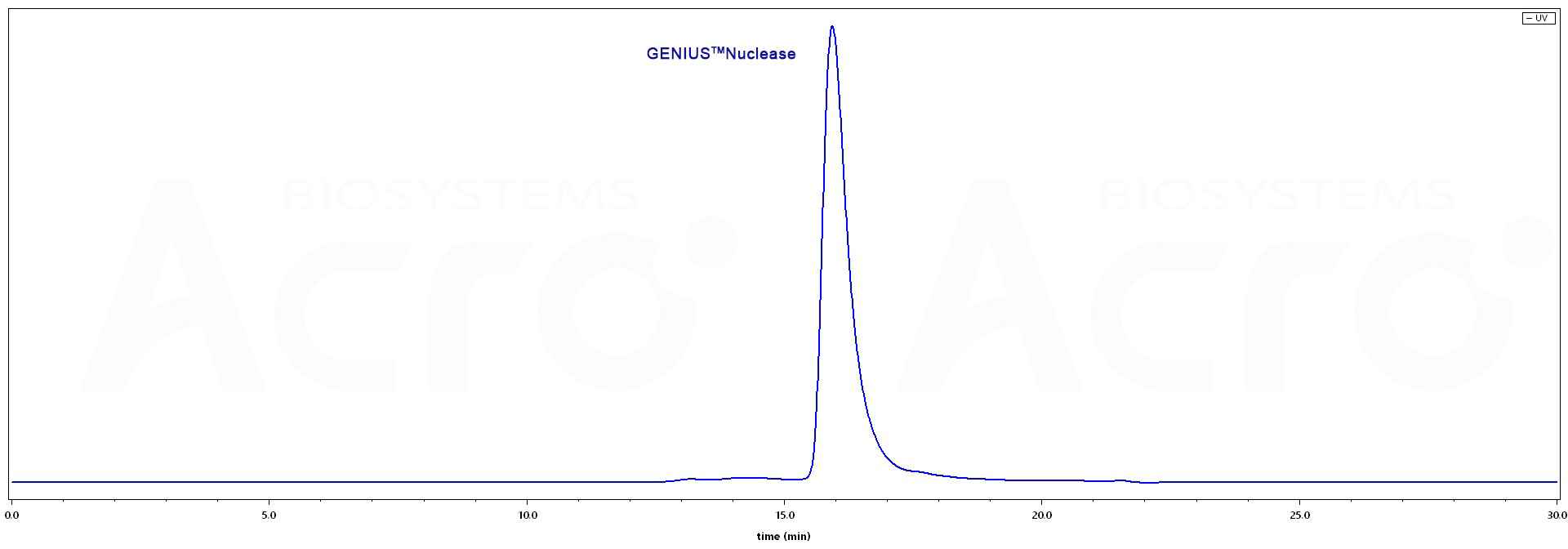
The purity of GENIUS™Nuclease (Cat. No. BEE-N3116) is more than 95% as determined by SEC-HPLC.
Report
Application example
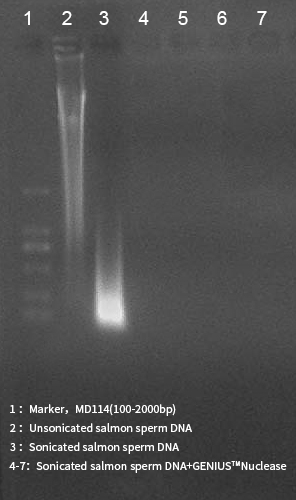
The result of GENIUS™Nuclease activity.























































 膜杰作
膜杰作 Star Staining
Star Staining







 +添加评论
+添加评论








